Research
Research in the Grant lab focuses on the development and application of state-of-the-art computational and theoretical techniques to investigate the structure, dynamics and interactions of biomedically important proteins.
At the interface of biochemistry, biophysics and bioinformatics the unique insight gained from our research is tightly coupled to a wide range of biochemical and biophysical experiments.
Using this approach we have recently developed a new class of allosteric Ras & Rho molecular switch inhibitors, reported the first rationally designed faster velocity kinesin motors, demonstrated for the first time that kinesin processivity can be modulated by rational mutation of the motor domain, discovered key atomistic detriments of allosteric activation in G proteins, and developed the Bio3D software package used by thousands of researchers around the world.
Our research goals are to advance these efforts to further facilitate the fields of protein design and drug discovery.
Molecular Motors & Molecular Switches
Deciphering the physical mechanisms underlying molecular motor and switch function and how their dysfunction is related to disease.
A major portion of our work asks about the common and divergent molecular mechanisms of kinesin motors and homologous G proteins switches. These mechanisms drive the internal coordination, communication and self-organization of eukaryotic cells; and their aberrant activities underlie a wide range of human cancers and neurodegenerative disorders.
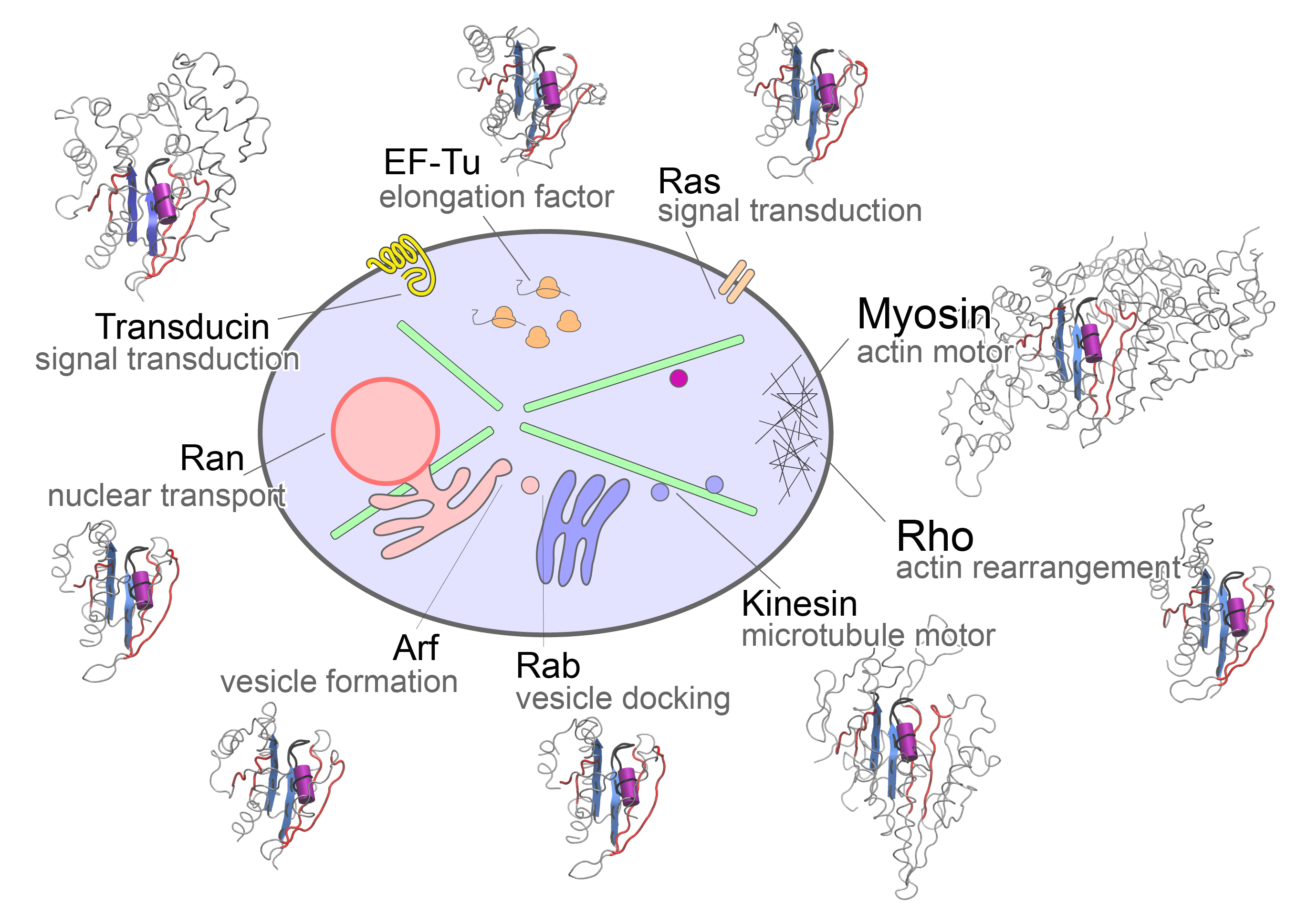
The molecular motors and switches that we focus on comprise a set of related nucleotide binding proteins that coordinate important cell processes including signaling, trafficking, transport, and protein synthesis. Our research aims to decipher the structural dynamic mechanisms by which these fascinating natural nanomachines function and how their dysfunction is related to disease.
Our recent findings on these systems include how to rationally engineer faster velocity longer running molecular motors and how to both allosterically activate and inhibit molecular switches that are malfunctioning in more than 20% of all human cancers.
Methods for Structural Bioinformatics & Computer Aided Drug Design
Integrating protein structural dynamics, evolutionary analysis & structure based drug discovery.
Proteins are structurally flexible molecules that often depend on their coordinated dynamic fluctuations to function properly. However, most current drug development strategies take one or a few rigid protein structures and design a single molecule to block its activity (i.e., catalytic pocket). We are developing a more sophisticated methodology that exploits flexible target structures, allosteric pocket identification and evolutionary analysis.
To this aim, our lab introduced Bio3D, a widely used open source R package for the analysis of protein sequence-structure-dynamics relationships. In particular, Bio3D is widely used by researchers around the world for combining information from all available experimental structures and sequences with that predicted by theory and computations.
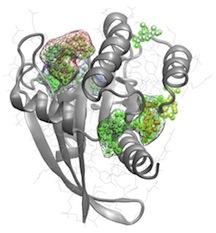 Motivated by the wide utility and use of Bio3D, together with our own research questions, we are currently working to extend functionality and interoperability with leading cheminformatics resources. This will enable the inclusion of protein structural flexibility and allosteric pocket detection into structure based drug design protocols.
Motivated by the wide utility and use of Bio3D, together with our own research questions, we are currently working to extend functionality and interoperability with leading cheminformatics resources. This will enable the inclusion of protein structural flexibility and allosteric pocket detection into structure based drug design protocols.
Merging these new methods in a single integrated environment represents an important advance that will greatly facilitate both bioinformatics research and the design of selective inhibitors.
Recent Publications
- Rapid Characterization of Allosteric Networks with Ensemble Normal Mode Analysis.
Yao, Skjærven, Grant. (2016)
J Phys Chem B. 2016 Apr 20. [Epub ahead of print]
( Journal | PubMed | PDF )
- Dynamic coupling and allosteric networks in the alpha subunit of heterotrimeric G proteins.
Yao, Malik, Griggs, Skjaerven, Traynor, Sivaramakrishnan, Grant. (2016)
J Biol Chem. 291(9), 4742-4753.
( Journal | PubMed | PDF )
- Mapping the processivity determinants of the kinesin-3 motor domain.
Scarabelli, Soppina, Yao, Atherton, Moores, Verhey, Grant. (2015)
Biophys J. 109, 1537–1540.
( Journal | PubMed | PDF )
- Enrichment of druggable conformations from apo protein structures using cosolvent-accelerated molecular dynamics.
Kalenkiewicz, Grant, Yang. (2015)
Biology. 4, 344-366
( Journal | PubMed | PDF )
- Integrating protein structural dynamics and evolutionary analysis with Bio3D.
Skjærven, Yao, Scarabelli, Grant. (2014)
BMC Bioinformatics. 15, 399
( Journal | PubMed | PDF )
- Kinesin-5 allosteric inhibitors uncouple the dynamics of nucleotide, microtubule and neck-linker binding sites.
Scarabelli, Grant. (2014)
Biophys J. 107, 2204–2213
( Journal | PubMed | PDF )
- Mapping the structural and dynamical features of kinesin motor domains.
Scarabelli, Grant. (2013)
PLoS Comput Biol. 9, e1003329
( Journal | PubMed | PDF )
- Domain opening and dynamic coupling in the alpha subunit of heterotrimeric G proteins.
Yao, Grant. (2013)
Biophys J. 105, L08–L10
( Journal | PubMed | PDF )
Access our complete publication list →
