BGGN-213, Lecture 17
Transcriptomics and the analysis of RNA-Seq data
Overiview
The data for this hands-on session comes from a published RNA-seq experiment where airway smooth muscle cells were treated with dexamethasone, a synthetic glucocorticoid steroid with anti-inflammatory effects (Himes et al. 2014).
Glucocorticoids are used, for example, by people with asthma to reduce inflammation of the airways. The anti-inflammatory effects on airway smooth muscle (ASM) cells has been known for some time but the underlying molecular mechanisms are unclear.
Himes et al. used RNA-seq to profile gene expression changes in four different ASM cell lines treated with dexamethasone glucocorticoid. They found a number of differentially expressed genes comparing dexamethasone-treated to control cells, but focus much of the discussion on a gene called CRISPLD2. This gene encodes a secreted protein known to be involved in lung development, and SNPs in this gene in previous GWAS studies are associated with inhaled corticosteroid resistance and bronchodilator response in asthma patients. They confirmed the upregulated CRISPLD2 mRNA expression with qPCR and increased protein expression using Western blotting.
In the experiment, four primary human ASM cell lines were treated with 1 micromolar dexamethasone for 18 hours. For each of the four cell lines, we have a treated and an untreated sample. They did their analysis using Tophat and Cufflinks similar to our Lecture 15 hands-on session. For a more detailed description of their analysis see the PubMed entry 24926665 and for raw data see the GEO entry GSE52778.
In this session we will read and explore the gene expression data from this experiment using base R functions and then perform a detailed analysis with the DESeq2 package from Bioconductor.
Section 1. Bioconductor and DESeq2 setup
As we already noted back in Lecture 10 and Lecture 11 Bioconductor is a large repository and resource for R packages that focus on analysis of high-throughput genomic data.
Bioconductor packages are installed differently than “regular”” R packages from CRAN. To install the core Bioconductor packages, copy and paste the following lines of code into your R console one at a time.
source("http://bioconductor.org/biocLite.R")
biocLite()
# For this class, you'll also need DESeq2:
biocLite("DESeq2")
The entire install process can take some time as there are many packages with dependencies on other packages. For some important notes on the install process please see https://bioboot.github.io/bggn213_f17//class-material/bioconductor_setup/. Your install process may produce some notes or other output. Generally, as long as you don’t get an error message, you’re good to move on.
Side-note: Aligning reads to a reference genome
The computational analysis of an RNA-seq experiment begins from the FASTQ files that contain the nucleotide sequence of each read and a quality score at each position. These reads must first be aligned to a reference genome or transcriptome. The output of this alignment step is commonly stored in a file format called SAM/BAM.
Once the reads have been aligned, there are a number of tools that can be used to count the number of reads/fragments that can be assigned to genomic features for each sample. These often take as input SAM/BAM alignment files and a file specifying the genomic features, e.g. a GFF3 or GTF file specifying the gene models as obtained from ENSEMBLE or UCSC.
In the workflow we’ll use here, the abundance of each transcript was quantified using kallisto (software, paper) and transcript-level abundance estimates were then summarized to the gene level to produce length-scaled counts using the R package txImport (software, paper), suitable for using in count-based analysis tools like DESeq. This is the starting point - a “count matrix”, where each cell indicates the number of reads mapping to a particular gene (in rows) for each sample (in columns).
Note: This is one of several well-established workflows for data pre-processing. The goal here is to provide a reference point to acquire fundamental skills with DESeq2 that will be applicable to other bioinformatics tools and workflows. In this regard, the following resources summarize a number of best practices for RNA-seq data analysis and pre-processing.
- Conesa, A. et al. “A survey of best practices for RNA-seq data analysis.” Genome Biology 17:13 (2016).
- Soneson, C., Love, M. I. & Robinson, M. D. “Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences.” F1000Res. 4:1521 (2016).
- Griffith, Malachi, et al. “Informatics for RNA sequencing: a web resource for analysis on the cloud.” PLoS Comput Biol 11.8: e1004393 (2015).
DESeq2 Required Inputs
As input, the DESeq2 package expects (1) a data.frame of count data (as obtained from RNA-seq or another high-throughput sequencing experiment) and (2) a second data.frame with information about the samples - often called sample metadata (or colData in DESeq2-speak because it supplies metadata/information about the columns of the countData matrix).

The “count matrix” (called the countData in DESeq2-speak) the value in the i-th row and the j-th column of the data.frame tells us how many reads can be assigned to gene i in sample j. Analogously, for other types of assays, the rows of this matrix might correspond e.g. to binding regions (with ChIP-Seq) or peptide sequences (with quantitative mass spectrometry).
For the sample metadata (called the colData in DESeq2-speak) samples are in rows and metadata about those samples are in columns. Notice that the first column of colData must match the column names of countData (except the first, which is the gene ID column).
Note from the DESeq2 vignette: The values in the input contData object should be counts of sequencing reads/fragments. This is important for DESeq2’s statistical model to hold, as only counts allow assessing the measurement precision correctly. It is important to never provide counts that were pre-normalized for sequencing depth/library size, as the statistical model is most powerful when applied to un-normalized counts, and is designed to account for library size differences internally.
Section 2. Import countData and colData into R
First, create a new RStudio project (File > New Project > New Directory > New Project) and download the input airway_scaledcounts.csv and airway_metadata.csv into a new data sub-directory of your project directory.
Begin a new R script and use the read.csv() function to read these count data and metadata files.
counts <- read.csv("data/airway_scaledcounts.csv", stringsAsFactors = FALSE)
metadata <- read.csv("data/airway_metadata.csv", stringsAsFactors = FALSE)
Now, take a look at each.
head(counts)
## ensgene SRR1039508 SRR1039509 SRR1039512 SRR1039513 SRR1039516
## 1 ENSG00000000003 723 486 904 445 1170
## 2 ENSG00000000005 0 0 0 0 0
## 3 ENSG00000000419 467 523 616 371 582
## 4 ENSG00000000457 347 258 364 237 318
## 5 ENSG00000000460 96 81 73 66 118
## 6 ENSG00000000938 0 0 1 0 2
## SRR1039517 SRR1039520 SRR1039521
## 1 1097 806 604
## 2 0 0 0
## 3 781 417 509
## 4 447 330 324
## 5 94 102 74
## 6 0 0 0
head(metadata)
## id dex celltype geo_id
## 1 SRR1039508 control N61311 GSM1275862
## 2 SRR1039509 treated N61311 GSM1275863
## 3 SRR1039512 control N052611 GSM1275866
## 4 SRR1039513 treated N052611 GSM1275867
## 5 SRR1039516 control N080611 GSM1275870
## 6 SRR1039517 treated N080611 GSM1275871
You can also use the View() function to view the entire object. Notice something here. The sample IDs in the metadata sheet (SRR1039508, SRR1039509, etc.) exactly match the column names of the countdata, except for the first column, which contains the Ensembl gene ID. This is important, and we’ll get more strict about it later on.
Section 3. Toy differential gene expression
Lets perform some exploratory differential gene expression analysis. Note: this analysis is for demonstration only. NEVER do differential expression analysis this way!
Look at the metadata object again to see which samples are control and which are drug treated
View(metadata)
If we look at our metadata, we see that the control samples are SRR1039508, SRR1039512, SRR1039516, and SRR1039520. This bit of code will first find the sample id for those labeled control. Then calculate the mean counts per gene across these samples:
control <- metadata[metadata[,"dex"]=="control",]
control.mean <- rowSums( counts[ ,control$id] )/4
names(control.mean) <- counts$ensgene
Q1. How would you make the above code more robust? What would happen if you were to add more samples. Would the values obtained with the excat code above be correct?
Q2. Follow the same procedure for the
treatedsamples (i.e. calculate the mean per gene accross drug treated samples and assign to a labeled vector calledtreated.mean)
We will combine our meancount data for bookkeeping purposes.
meancounts <- data.frame(control.mean, treated.mean)
Directly comparing the raw counts is going to be problematic if we just happened to sequence one group at a higher depth than another. Later on we’ll do this analysis properly, normalizing by sequencing depth per sample using a better approach. But for now, colSums() the data to show the sum of the mean counts across all genes for each group. Your answer should look like this:
## control.mean treated.mean
## 23005324 22196524
Q3. Create a scatter plot showing the mean of the treated samples against the mean of the control samples. Your plot should look something like the following.
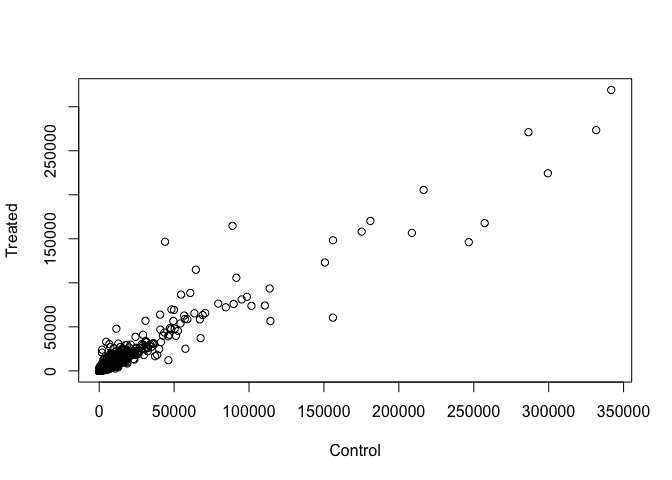
Wait a sec. There are 60,000-some rows in this data, but I’m only seeing a few dozen dots at most outside of the big clump around the origin. Try plotting both axes on a log scale (hint: see the help for ?plot.default to see how to set log axis.
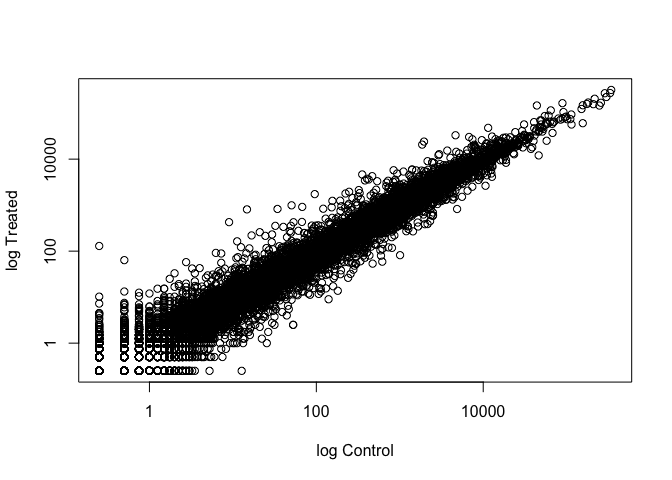
We can find candidate differentially expressed genes by looking for genes with a large change between control and dex-treated samples. We usually look at the log2 of the fold change, because this has better mathematical properties.
Here we calculate log2foldchange, add it to our meancounts data.frame and inspect the results either with the head() or the View() function for example.
meancounts$log2fc <- log2(meancounts[,"treated.mean"]/meancounts[,"control.mean"])
head(meancounts)
## control.mean treated.mean log2fc
## ENSG00000000003 900.75 658.00 -0.45303916
## ENSG00000000005 0.00 0.00 NaN
## ENSG00000000419 520.50 546.00 0.06900279
## ENSG00000000457 339.75 316.50 -0.10226805
## ENSG00000000460 97.25 78.75 -0.30441833
## ENSG00000000938 0.75 0.00 -Inf
There are a couple of “weird” results. Namely, the NaN (“not a number””) and -Inf (negative infinity) results.
The NaN is returned when you divide by zero and try to take the log. The -Inf is returned when you try to take the log of zero. It turns out that there are a lot of genes with zero expression. Let’s filter our data to remove these genes. Again inspect your result (and the intermediate steps) to see if things make sense to you
zero.vals <- which(meancounts[,1:2]==0, arr.ind=TRUE)
to.rm <- unique(zero.vals[,1])
mycounts <- meancounts[-to.rm,]
head(mycounts)
## control.mean treated.mean log2fc
## ENSG00000000003 900.75 658.00 -0.45303916
## ENSG00000000419 520.50 546.00 0.06900279
## ENSG00000000457 339.75 316.50 -0.10226805
## ENSG00000000460 97.25 78.75 -0.30441833
## ENSG00000000971 5219.00 6687.50 0.35769358
## ENSG00000001036 2327.00 1785.75 -0.38194109
Q4. What is the purpose of the
arr.indargument in the which() function call above? Why would we then take the first column of the output and need to call the unique() function?
A common threshold used for calling something differentially expressed is a log2(FoldChange) of greater than 2 or less than -2. Let’s filter the dataset both ways to see how many genes are up or down-regulated.
up.ind <- mycounts$log2fc > 2
down.ind <- mycounts$log2fc < (-2)
Q5. Using the
up.indanddown.indvectors above can you determine how many up and down regulated genes we have at the greater than 2 fc level?
## [1] "Up: 250"
## [1] "Down: 367"
In total, you should of reported 617 differentially expressed genes, in either direction.
Section 4. Adding annotation data
Our mycounts result table so far only contains the Ensembl gene IDs. However, alternative gene names and extra annotation are usually required for informative for interpretation.
We can add annotation from a supplied CSV file, such as those available from ENSEMBLE or UCSC. The annotables_grch38.csv annotation table links the unambiguous Ensembl gene ID to other useful annotation like the gene symbol, full gene name, location, Entrez gene ID, etc.
anno <- read.csv("data/annotables_grch38.csv")
head(anno)
## ensgene entrez symbol chr start end strand
## 1 ENSG00000000003 7105 TSPAN6 X 100627109 100639991 -1
## 2 ENSG00000000005 64102 TNMD X 100584802 100599885 1
## 3 ENSG00000000419 8813 DPM1 20 50934867 50958555 -1
## 4 ENSG00000000457 57147 SCYL3 1 169849631 169894267 -1
## 5 ENSG00000000460 55732 C1orf112 1 169662007 169854080 1
## 6 ENSG00000000938 2268 FGR 1 27612064 27635277 -1
## biotype
## 1 protein_coding
## 2 protein_coding
## 3 protein_coding
## 4 protein_coding
## 5 protein_coding
## 6 protein_coding
## description
## 1 tetraspanin 6 [Source:HGNC Symbol;Acc:HGNC:11858]
## 2 tenomodulin [Source:HGNC Symbol;Acc:HGNC:17757]
## 3 dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit [Source:HGNC Symbol;Acc:HGNC:3005]
## 4 SCY1-like, kinase-like 3 [Source:HGNC Symbol;Acc:HGNC:19285]
## 5 chromosome 1 open reading frame 112 [Source:HGNC Symbol;Acc:HGNC:25565]
## 6 FGR proto-oncogene, Src family tyrosine kinase [Source:HGNC Symbol;Acc:HGNC:3697]
Ideally we want this annotation data mapped (or merged) with our mycounts data. In a previous class on writing R functions we introduced the merge() function, which is one common way to do this.
Q6. From consulting the help page for the merge() function can you set the
by.xandby.yarguments appropriately to annotate ourmycountsdata.frame with all the available annotation data in yourannodata.frame?
Examine your results with the View() function. It should look something like the following:
## Row.names control.mean treated.mean log2fc entrez symbol
## 1 ENSG00000000003 900.75 658.00 -0.45303916 7105 TSPAN6
## 2 ENSG00000000419 520.50 546.00 0.06900279 8813 DPM1
## 3 ENSG00000000457 339.75 316.50 -0.10226805 57147 SCYL3
## 4 ENSG00000000460 97.25 78.75 -0.30441833 55732 C1orf112
## 5 ENSG00000000971 5219.00 6687.50 0.35769358 3075 CFH
## 6 ENSG00000001036 2327.00 1785.75 -0.38194109 2519 FUCA2
## chr start end strand biotype
## 1 X 100627109 100639991 -1 protein_coding
## 2 20 50934867 50958555 -1 protein_coding
## 3 1 169849631 169894267 -1 protein_coding
## 4 1 169662007 169854080 1 protein_coding
## 5 1 196651878 196747504 1 protein_coding
## 6 6 143494811 143511690 -1 protein_coding
## description
## 1 tetraspanin 6 [Source:HGNC Symbol;Acc:HGNC:11858]
## 2 dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit [Source:HGNC Symbol;Acc:HGNC:3005]
## 3 SCY1-like, kinase-like 3 [Source:HGNC Symbol;Acc:HGNC:19285]
## 4 chromosome 1 open reading frame 112 [Source:HGNC Symbol;Acc:HGNC:25565]
## 5 complement factor H [Source:HGNC Symbol;Acc:HGNC:4883]
## 6 fucosidase, alpha-L- 2, plasma [Source:HGNC Symbol;Acc:HGNC:4008]
In cases where you don’t have a preferred annotation file at hand you can use other Bioconductor packages for annotation.
Bioconductor’s annotation packages help with mapping various ID schemes to each other. Here we load the AnnotationDbi package and the annotation package org.Hs.eg.db.
library("AnnotationDbi")
library("org.Hs.eg.db")
Note: You may have to install these with the
biocLite("AnnotationDbi")function etc.
This is the organism annotation package (“org”) for Homo sapiens (“Hs”), organized as an AnnotationDbi database package (“db”), using Entrez Gene IDs (“eg”) as primary key. To get a list of all available key types, use:
columns(org.Hs.eg.db)
## [1] "ACCNUM" "ALIAS" "ENSEMBL" "ENSEMBLPROT"
## [5] "ENSEMBLTRANS" "ENTREZID" "ENZYME" "EVIDENCE"
## [9] "EVIDENCEALL" "GENENAME" "GO" "GOALL"
## [13] "IPI" "MAP" "OMIM" "ONTOLOGY"
## [17] "ONTOLOGYALL" "PATH" "PFAM" "PMID"
## [21] "PROSITE" "REFSEQ" "SYMBOL" "UCSCKG"
## [25] "UNIGENE" "UNIPROT"
We can use the mapIds() function to add individual columns to our results table. We provide the row names of our results table as a key, and specify that keytype=ENSEMBL. The column argument tells the mapIds() function which information we want, and the multiVals argument tells the function what to do if there are multiple possible values for a single input value. Here we ask to just give us back the first one that occurs in the database.
mycounts$symbol <- mapIds(org.Hs.eg.db,
keys=row.names(mycounts),
column="SYMBOL",
keytype="ENSEMBL",
multiVals="first")
## 'select()' returned 1:many mapping between keys and columns
Q7. Run the mapIds() function two more times to add the Entrez ID and UniProt accession as new columns called
mycounts$entrezandmycounts$uniprot. The head() of your results should look like the following:
## 'select()' returned 1:many mapping between keys and columns
## 'select()' returned 1:many mapping between keys and columns
## control.mean treated.mean log2fc symbol entrez
## ENSG00000000003 900.75 658.00 -0.45303916 TSPAN6 7105
## ENSG00000000419 520.50 546.00 0.06900279 DPM1 8813
## ENSG00000000457 339.75 316.50 -0.10226805 SCYL3 57147
## ENSG00000000460 97.25 78.75 -0.30441833 C1orf112 55732
## ENSG00000000971 5219.00 6687.50 0.35769358 CFH 3075
## ENSG00000001036 2327.00 1785.75 -0.38194109 FUCA2 2519
## uniprot
## ENSG00000000003 A0A024RCI0
## ENSG00000000419 O60762
## ENSG00000000457 Q8IZE3
## ENSG00000000460 A0A024R922
## ENSG00000000971 A0A024R962
## ENSG00000001036 Q9BTY2
Q8. Examine your annotated results for those genes with a log2(FoldChange) of greater than 2 (or less than -2 if you prefer) with the View() function. What do you notice? Would you trust these results? Why or why not?
head(mycounts[up.ind,])
## control.mean treated.mean log2fc symbol entrez
## ENSG00000004799 270.50 1429.25 2.401558 PDK4 5166
## ENSG00000006788 2.75 19.75 2.844349 MYH13 8735
## ENSG00000008438 0.50 2.75 2.459432 PGLYRP1 8993
## ENSG00000011677 0.50 2.25 2.169925 GABRA3 2556
## ENSG00000015413 0.50 3.00 2.584963 DPEP1 1800
## ENSG00000015592 0.50 2.25 2.169925 STMN4 81551
## uniprot
## ENSG00000004799 A4D1H4
## ENSG00000006788 Q9UKX3
## ENSG00000008438 O75594
## ENSG00000011677 P34903
## ENSG00000015413 A0A140VJI3
## ENSG00000015592 Q9H169
Section 5. DESeq2 analysis
Let’s do this the right way. DESeq2 is an R package for analyzing count-based NGS data like RNA-seq. It is available from Bioconductor. Bioconductor is a project to provide tools for analyzing high-throughput genomic data including RNA-seq, ChIP-seq and arrays. You can explore Bioconductor packages here.
Bioconductor packages usually have great documentation in the form of vignettes. For a great example, take a look at the DESeq2 vignette for analyzing count data. This 40+ page manual is packed full of examples on using DESeq2, importing data, fitting models, creating visualizations, references, etc.
Just like R packages from CRAN, you only need to install Bioconductor packages once (instructions here), then load them every time you start a new R session.
library(DESeq2)
citation("DESeq2")
Take a second and read through all the stuff that flies by the screen when you load the DESeq2 package. When you first installed DESeq2 it may have taken a while, because DESeq2 depends on a number of other R packages (S4Vectors, BiocGenerics, parallel, IRanges, etc.) Each of these, in turn, may depend on other packages. These are all loaded into your working environment when you load DESeq2. Also notice the lines that start with The following objects are masked from 'package:....
Importing data
Bioconductor software packages often define and use custom class objects for storing data. This helps to ensure that all the needed data for analysis (and the results) are available. DESeq works on a particular type of object called a DESeqDataSet. The DESeqDataSet is a single object that contains input values, intermediate calculations like how things are normalized, and all results of a differential expression analysis.
You can construct a DESeqDataSet from (1) a count matrix, (2) a metadata file, and (3) a formula indicating the design of the experiment.
We have talked about (1) and (2) previously. The third needed item that has to be specified at the beginning of the analysis is a design formula. This tells DESeq2 which columns in the sample information table (colData) specify the experimental design (i.e. which groups the samples belong to) and how these factors should be used in the analysis. Essentially, this formula expresses how the counts for each gene depend on the variables in colData.
Take a look at metadata again. The thing we’re interested in is the dex column, which tells us which samples are treated with dexamethasone versus which samples are untreated controls. We’ll specify the design with a tilde, like this: design=~dex. (The tilde is the shifted key to the left of the number 1 key on my keyboard. It looks like a little squiggly line).
We will use the DESeqDataSetFromMatrix() function to build the required DESeqDataSet object and call it dds, short for our DESeqDataSet. If you get a warning about “some variables in design formula are characters, converting to factors” don’t worry about it. Take a look at the dds object once you create it.
dds <- DESeqDataSetFromMatrix(countData=counts,
colData=metadata,
design=~dex,
tidy=TRUE)
dds
## class: DESeqDataSet
## dim: 38694 8
## metadata(1): version
## assays(1): counts
## rownames(38694): ENSG00000000003 ENSG00000000005 ...
## ENSG00000283120 ENSG00000283123
## rowData names(0):
## colnames(8): SRR1039508 SRR1039509 ... SRR1039520 SRR1039521
## colData names(4): id dex celltype geo_id
DESeq pipeline
Next, let’s run the DESeq pipeline on the dataset, and reassign the resulting object back to the same variable. Before we start, dds is a bare-bones DESeqDataSet. The DESeq() function takes a DESeqDataSet and returns a DESeqDataSet, but with lots of other information filled in (normalization, dispersion estimates, differential expression results, etc). Notice how if we try to access these objects before running the analysis, nothing exists.
sizeFactors(dds)
## NULL
dispersions(dds)
## NULL
results(dds)
## Error in results(dds): couldn't find results. you should first run DESeq()
Here, we’re running the DESeq pipeline on the dds object, and reassigning the whole thing back to dds, which will now be a DESeqDataSet populated with all those values. Get some help on ?DESeq (notice, no “2” on the end). This function calls a number of other functions within the package to essentially run the entire pipeline (normalizing by library size by estimating the “size factors,” estimating dispersion for the negative binomial model, and fitting models and getting statistics for each gene for the design specified when you imported the data).
dds <- DESeq(dds)
## estimating size factors
## estimating dispersions
## gene-wise dispersion estimates
## mean-dispersion relationship
## final dispersion estimates
## fitting model and testing
Getting results
Since we’ve got a fairly simple design (single factor, two groups, treated versus control), we can get results out of the object simply by calling the results() function on the DESeqDataSet that has been run through the pipeline. The help page for ?results and the vignette both have extensive documentation about how to pull out the results for more complicated models (multi-factor experiments, specific contrasts, interaction terms, time courses, etc.).
res <- results(dds)
res
## log2 fold change (MLE): dex treated vs control
## Wald test p-value: dex treated vs control
## DataFrame with 38694 rows and 6 columns
## baseMean log2FoldChange lfcSE stat pvalue
## <numeric> <numeric> <numeric> <numeric> <numeric>
## ENSG00000000003 747.19420 -0.35070283 0.1682342 -2.0846111 0.03710462
## ENSG00000000005 0.00000 NA NA NA NA
## ENSG00000000419 520.13416 0.20610652 0.1010134 2.0403876 0.04131173
## ENSG00000000457 322.66484 0.02452714 0.1451103 0.1690242 0.86577762
## ENSG00000000460 87.68263 -0.14714409 0.2569657 -0.5726216 0.56690095
## ... ... ... ... ... ...
## ENSG00000283115 0.000000 NA NA NA NA
## ENSG00000283116 0.000000 NA NA NA NA
## ENSG00000283119 0.000000 NA NA NA NA
## ENSG00000283120 0.974916 -0.6682308 1.694063 -0.3944544 0.6932456
## ENSG00000283123 0.000000 NA NA NA NA
## padj
## <numeric>
## ENSG00000000003 0.1630257
## ENSG00000000005 NA
## ENSG00000000419 0.1757326
## ENSG00000000457 0.9616577
## ENSG00000000460 0.8157061
## ... ...
## ENSG00000283115 NA
## ENSG00000283116 NA
## ENSG00000283119 NA
## ENSG00000283120 NA
## ENSG00000283123 NA
Either click on the res object in the environment pane or pass it to View() to bring it up in a data viewer. Why do you think so many of the adjusted p-values are missing (NA)? Try looking at the baseMean column, which tells you the average overall expression of this gene, and how that relates to whether or not the p-value was missing. Go to the DESeq2 vignette and read the section about “Independent filtering and multiple testing.”
Note. The goal of independent filtering is to filter out those tests from the procedure that have no, or little chance of showing significant evidence, without even looking at the statistical result. Genes with very low counts are not likely to see significant differences typically due to high dispersion. This results in increased detection power at the same experiment-wide type I error [i.e., better FDRs].
We can summarize some basic tallies using the summary function.
summary(res)
##
## out of 25258 with nonzero total read count
## adjusted p-value < 0.1
## LFC > 0 (up) : 1564, 6.2%
## LFC < 0 (down) : 1188, 4.7%
## outliers [1] : 142, 0.56%
## low counts [2] : 9971, 39%
## (mean count < 10)
## [1] see 'cooksCutoff' argument of ?results
## [2] see 'independentFiltering' argument of ?results
We can order our results table by the smallest p value:
resOrdered <- res[order(res$pvalue),]
The results function contains a number of arguments to customize the results table. By default the argument alpha is set to 0.1. If the adjusted p value cutoff will be a value other than 0.1, alpha should be set to that value:
res05 <- results(dds, alpha=0.05)
summary(res05)
##
## out of 25258 with nonzero total read count
## adjusted p-value < 0.05
## LFC > 0 (up) : 1237, 4.9%
## LFC < 0 (down) : 933, 3.7%
## outliers [1] : 142, 0.56%
## low counts [2] : 9033, 36%
## (mean count < 6)
## [1] see 'cooksCutoff' argument of ?results
## [2] see 'independentFiltering' argument of ?results
The more generic way to access the actual subset of the data.frame passing a threshold like this is with the subset() function, e.g.:
resSig05 <- subset(as.data.frame(res), padj < 0.05)
nrow(resSig05)
## [1] 2182
Q9. How many are significant with an adjusted p-value < 0.05? How about 0.01? Save this last set of results as
resSig01.
Q10. Using either the previously generated
annoobject (annotations from the fileannotables_grch38.csvfile) or the mapIds() function (from the AnnotationDbi package) add annotation to yourres01results data.frame.
## [1] 1437
You can arrange and view the results by the adjusted p-value
ord <- order( resSig01$padj )
#View(res01[ord,])
head(resSig01[ord,])
## baseMean log2FoldChange lfcSE stat
## ENSG00000152583 954.7709 4.368359 0.23713648 18.42129
## ENSG00000179094 743.2527 2.863888 0.17555825 16.31304
## ENSG00000116584 2277.9135 -1.034700 0.06505273 -15.90556
## ENSG00000189221 2383.7537 3.341544 0.21241508 15.73120
## ENSG00000120129 3440.7038 2.965211 0.20370277 14.55656
## ENSG00000148175 13493.9204 1.427168 0.10036663 14.21955
## pvalue padj symbol
## ENSG00000152583 8.867079e-76 1.342919e-71 SPARCL1
## ENSG00000179094 7.972621e-60 6.037267e-56 PER1
## ENSG00000116584 5.798513e-57 2.927283e-53 ARHGEF2
## ENSG00000189221 9.244206e-56 3.500088e-52 MAOA
## ENSG00000120129 5.306416e-48 1.607313e-44 DUSP1
## ENSG00000148175 6.929711e-46 1.749175e-42 STOM
Finally, let’s write out the ordered significant results with annotations. See the help for ?write.csv if you are unsure here.
write.csv(resSig01[ord,], "signif01_results.csv")
Section 6. Data Visualization
Plotting counts
DESeq2 offers a function called plotCounts() that takes a DESeqDataSet that has been run through the pipeline, the name of a gene, and the name of the variable in the colData that you’re interested in, and plots those values. See the help for ?plotCounts. Let’s first see what the gene ID is for the CRISPLD2 gene using:
i <- grep("CRISPLD2", resSig01$symbol)
resSig01[i,]
## baseMean log2FoldChange lfcSE stat pvalue
## ENSG00000103196 3096.159 2.626034 0.2674705 9.818031 9.416441e-23
## padj symbol
## ENSG00000103196 3.395524e-20 CRISPLD2
rownames(resSig01[i,])
## [1] "ENSG00000103196"
Now, with that gene ID in hand let’s plot the counts, where our intgroup, or “interesting group” variable is the “dex” column.
plotCounts(dds, gene="ENSG00000103196", intgroup="dex")
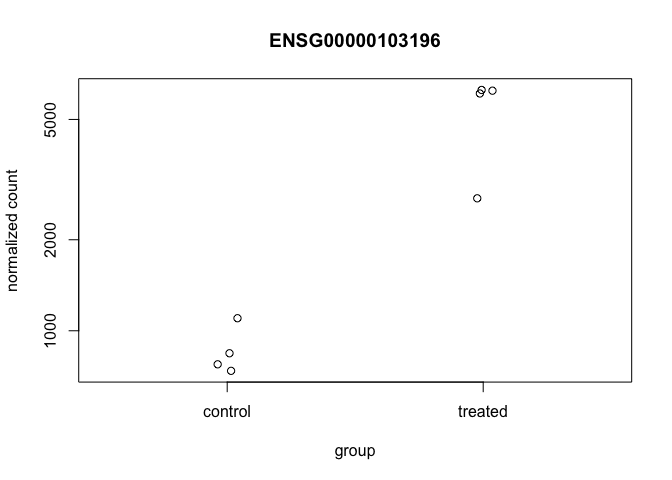
That’s just okay. Keep looking at the help for ?plotCounts. Notice that we could have actually returned the data instead of plotting. We could then pipe this to ggplot and make our own figure. Let’s make a boxplot.
# Return the data
d <- plotCounts(dds, gene="ENSG00000103196", intgroup="dex", returnData=TRUE)
This returned object is a data.frame all setup for ggplot2 based plotting. For example:
library(ggplot2)
ggplot(d, aes(dex, count)) + geom_boxplot(aes(fill=dex)) + scale_y_log10() + ggtitle("CRISPLD2")
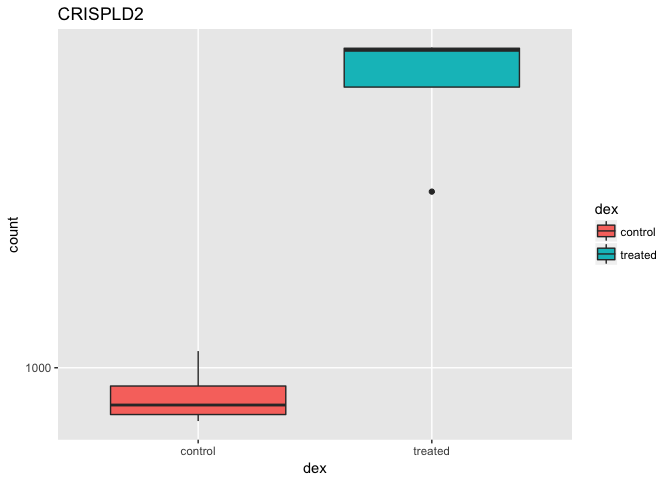
You can also plot this with base graphics and the boxplot() function if you prefer.
MA & Volcano plots
Let’s make some commonly produced visualizations from this data. First, let’s add a column called sig to our full res results that evaluates to TRUE if padj<0.05, and FALSE if not, and NA if padj is also NA.
res$sig <- res$padj<0.05
# How many of each?
table(res$sig)
##
## FALSE TRUE
## 12963 2182
sum(is.na(res$sig))
## [1] 23549
Look up the Wikipedia articles on MA plots and volcano plots. An MA plot shows the average expression on the X-axis and the log fold change on the y-axis. A volcano plot shows the log fold change on the X-axis, and the −log10 of the p-value on the Y-axis (the more significant the p-value, the larger the −log10 of that value will be).
## Warning: Transformation introduced infinite values in continuous x-axis
## Warning: Removed 13436 rows containing missing values (geom_point).
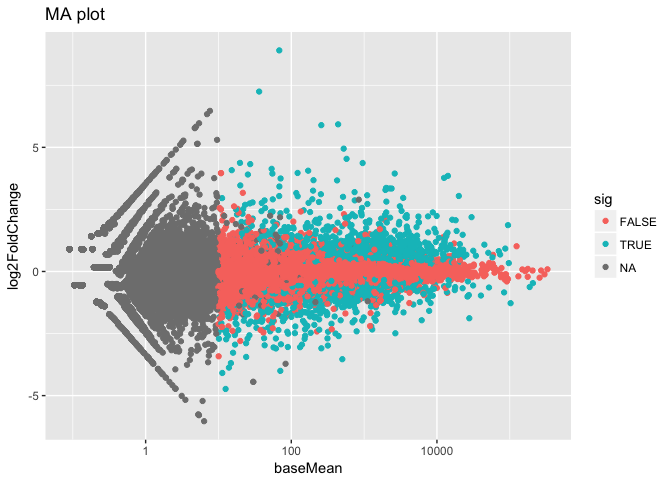
In built MA-plot
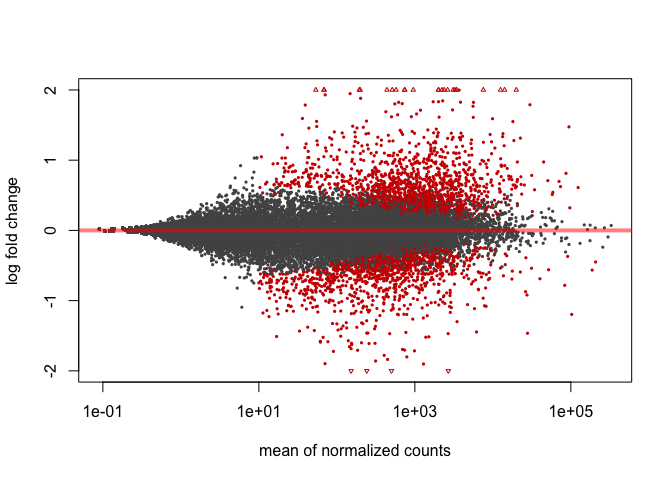
In DESeq2, the function plotMA() shows the log2 fold changes attributable to a given variable over the mean of normalized counts for all the samples in the DESeqDataSet. Points will be colored red if the adjusted p value is less than 0.1. Points which fall out of the window are plotted as open triangles pointing either up or down.
plotMA(res, ylim=c(-2,2))
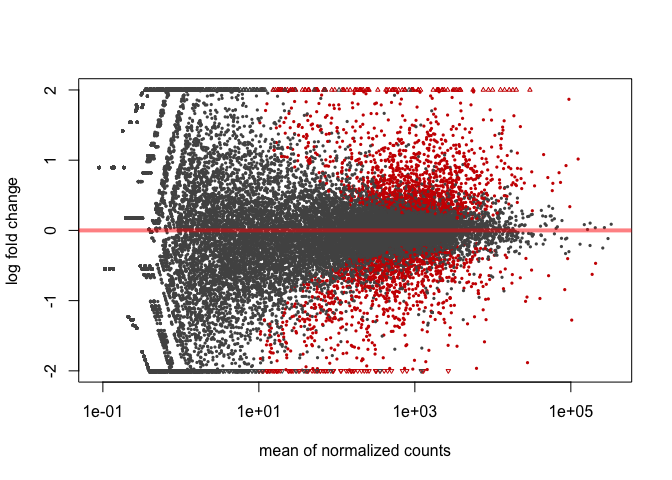
It is often more useful to visualize the MA-plot for so-called shrunken log2 fold changes, which remove the noise associated with log2 fold changes from low count genes.
resLFC <- lfcShrink(dds, coef=2)
resLFC
## log2 fold change (MAP): dex treated vs control
## Wald test p-value: dex treated vs control
## DataFrame with 38694 rows and 6 columns
## baseMean log2FoldChange lfcSE stat pvalue
## <numeric> <numeric> <numeric> <numeric> <numeric>
## ENSG00000000003 747.19420 -0.31838595 0.15271739 -2.0846111 0.03710462
## ENSG00000000005 0.00000 NA NA NA NA
## ENSG00000000419 520.13416 0.19883048 0.09744556 2.0403876 0.04131173
## ENSG00000000457 322.66484 0.02280238 0.13491699 0.1690242 0.86577762
## ENSG00000000460 87.68263 -0.11887370 0.20772938 -0.5726216 0.56690095
## ... ... ... ... ... ...
## ENSG00000283115 0.000000 NA NA NA NA
## ENSG00000283116 0.000000 NA NA NA NA
## ENSG00000283119 0.000000 NA NA NA NA
## ENSG00000283120 0.974916 -0.05944174 0.1514839 -0.3944544 0.6932456
## ENSG00000283123 0.000000 NA NA NA NA
## padj
## <numeric>
## ENSG00000000003 0.1630257
## ENSG00000000005 NA
## ENSG00000000419 0.1757326
## ENSG00000000457 0.9616577
## ENSG00000000460 0.8157061
## ... ...
## ENSG00000283115 NA
## ENSG00000283116 NA
## ENSG00000283119 NA
## ENSG00000283120 NA
## ENSG00000283123 NA
plotMA(resLFC, ylim=c(-2,2))
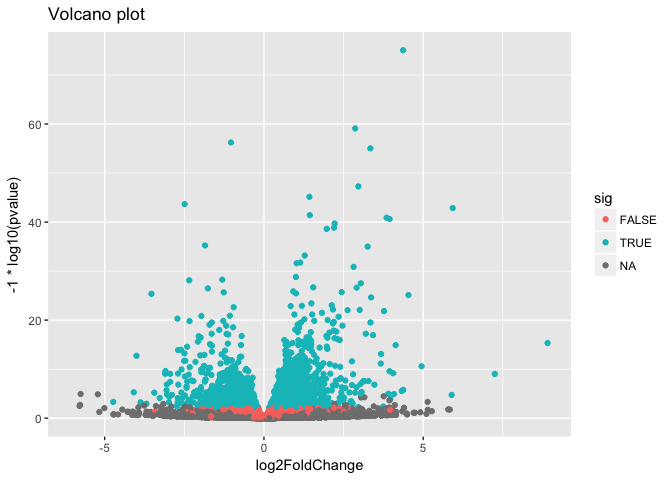
Volcano plot
Make a volcano plot. Similarly, color-code by whether it’s significant or not.
ggplot(as.data.frame(res), aes(log2FoldChange, -1*log10(pvalue), col=sig)) +
geom_point() +
ggtitle("Volcano plot")
## Warning: Removed 13578 rows containing missing values (geom_point).
Section 7. Side-note: Transformation
To test for differential expression we operate on raw counts. But for other downstream analyses like heatmaps, PCA, or clustering, we need to work with transformed versions of the data, because it’s not clear how to best compute a distance metric on untransformed counts. The go-to choice might be a log transformation. But because many samples have a zero count (and log(0)= − ∞, you might try using pseudocounts, i. e. y = log(n + 1) or more generally, y = log(n + n0), where n represents the count values and n0 is some positive constant.
But there are other approaches that offer better theoretical justification and a rational way of choosing the parameter equivalent to n0, and they produce transformed data on the log scale that’s normalized to library size. One is called a variance stabilizing transformation (VST), and it also removes the dependence of the variance on the mean, particularly the high variance of the log counts when the mean is low.
vsdata <- vst(dds, blind=FALSE)
PCA
Let’s do some exploratory plotting of the data using principal components analysis on the variance stabilized data from above. Let’s use the DESeq2-provided plotPCA function. See the help for ?plotPCA and notice that it also has a returnData option, just like plotCounts.
plotPCA(vsdata, intgroup="dex")
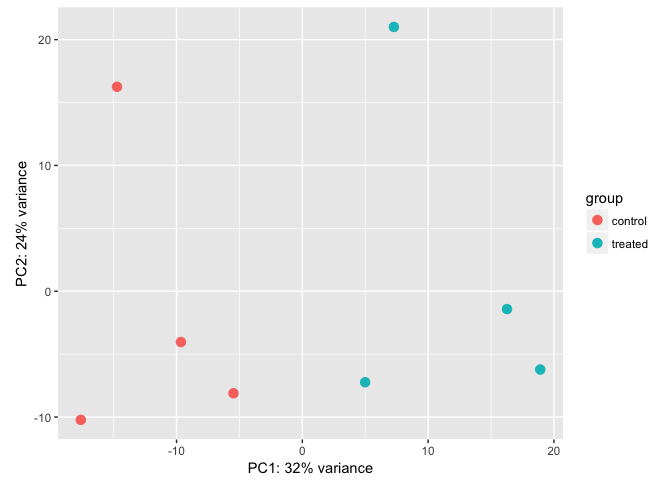
Principal Components Analysis (PCA) is a dimension reduction and visualization technique that is here used to project the multivariate data vector of each sample into a two-dimensional plot, such that the spatial arrangement of the points in the plot reflects the overall data (dis)similarity between the samples. In essence, principal component analysis distills all the global variation between samples down to a few variables called principal components. The majority of variation between the samples can be summarized by the first principal component, which is shown on the x-axis. The second principal component summarizes the residual variation that isn’t explained by PC1. PC2 is shown on the y-axis. The percentage of the global variation explained by each principal component is given in the axis labels. In a two-condition scenario (e.g., mutant vs WT, or treated vs control), you might expect PC1 to separate the two experimental conditions, so for example, having all the controls on the left and all experimental samples on the right (or vice versa - the units and directionality isn’t important). The secondary axis may separate other aspects of the design - cell line, time point, etc. Very often the experimental design is reflected in the PCA plot, and in this case, it is. But this kind of diagnostic can be useful for finding outliers, investigating batch effects, finding sample swaps, and other technical problems with the data. This YouTube video from the Genetics Department at UNC gives a very accessible explanation of what PCA is all about in the context of a gene expression experiment, without the need for an advanced math background. Take a look. s
Session Information
The sessionInfo() prints version information about R and any attached packages. It’s a good practice to always run this command at the end of your R session and record it for the sake of reproducibility in the future.
sessionInfo()
## R version 3.4.1 (2017-06-30)
## Platform: x86_64-apple-darwin15.6.0 (64-bit)
## Running under: macOS Sierra 10.12.6
##
## Matrix products: default
## BLAS: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRblas.0.dylib
## LAPACK: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRlapack.dylib
##
## locale:
## [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
##
## attached base packages:
## [1] parallel stats4 stats graphics grDevices utils datasets
## [8] methods base
##
## other attached packages:
## [1] ggplot2_2.2.1 DESeq2_1.18.1
## [3] SummarizedExperiment_1.8.0 DelayedArray_0.4.1
## [5] matrixStats_0.52.2 GenomicRanges_1.30.0
## [7] GenomeInfoDb_1.14.0 org.Hs.eg.db_3.5.0
## [9] AnnotationDbi_1.40.0 IRanges_2.12.0
## [11] S4Vectors_0.16.0 Biobase_2.38.0
## [13] BiocGenerics_0.24.0
##
## loaded via a namespace (and not attached):
## [1] locfit_1.5-9.1 Rcpp_0.12.13
## [3] lattice_0.20-35 rprojroot_1.2
## [5] digest_0.6.12 plyr_1.8.4
## [7] backports_1.1.1 acepack_1.4.1
## [9] RSQLite_2.0 evaluate_0.10.1
## [11] zlibbioc_1.24.0 rlang_0.1.4
## [13] lazyeval_0.2.1 data.table_1.10.4-3
## [15] annotate_1.56.1 blob_1.1.0
## [17] rpart_4.1-11 Matrix_1.2-12
## [19] checkmate_1.8.5 rmarkdown_1.8
## [21] labeling_0.3 splines_3.4.1
## [23] BiocParallel_1.12.0 geneplotter_1.56.0
## [25] stringr_1.2.0 foreign_0.8-69
## [27] htmlwidgets_0.9 RCurl_1.95-4.8
## [29] bit_1.1-12 munsell_0.4.3
## [31] compiler_3.4.1 pkgconfig_2.0.1
## [33] base64enc_0.1-3 htmltools_0.3.6
## [35] nnet_7.3-12 tibble_1.3.4
## [37] gridExtra_2.3 htmlTable_1.9
## [39] GenomeInfoDbData_0.99.1 Hmisc_4.0-3
## [41] XML_3.98-1.9 bitops_1.0-6
## [43] grid_3.4.1 xtable_1.8-2
## [45] gtable_0.2.0 DBI_0.7
## [47] magrittr_1.5 scales_0.5.0
## [49] stringi_1.1.6 XVector_0.18.0
## [51] genefilter_1.60.0 latticeExtra_0.6-28
## [53] Formula_1.2-2 RColorBrewer_1.1-2
## [55] tools_3.4.1 bit64_0.9-7
## [57] survival_2.41-3 yaml_2.1.14
## [59] colorspace_1.3-2 cluster_2.0.6
## [61] memoise_1.1.0 knitr_1.17
