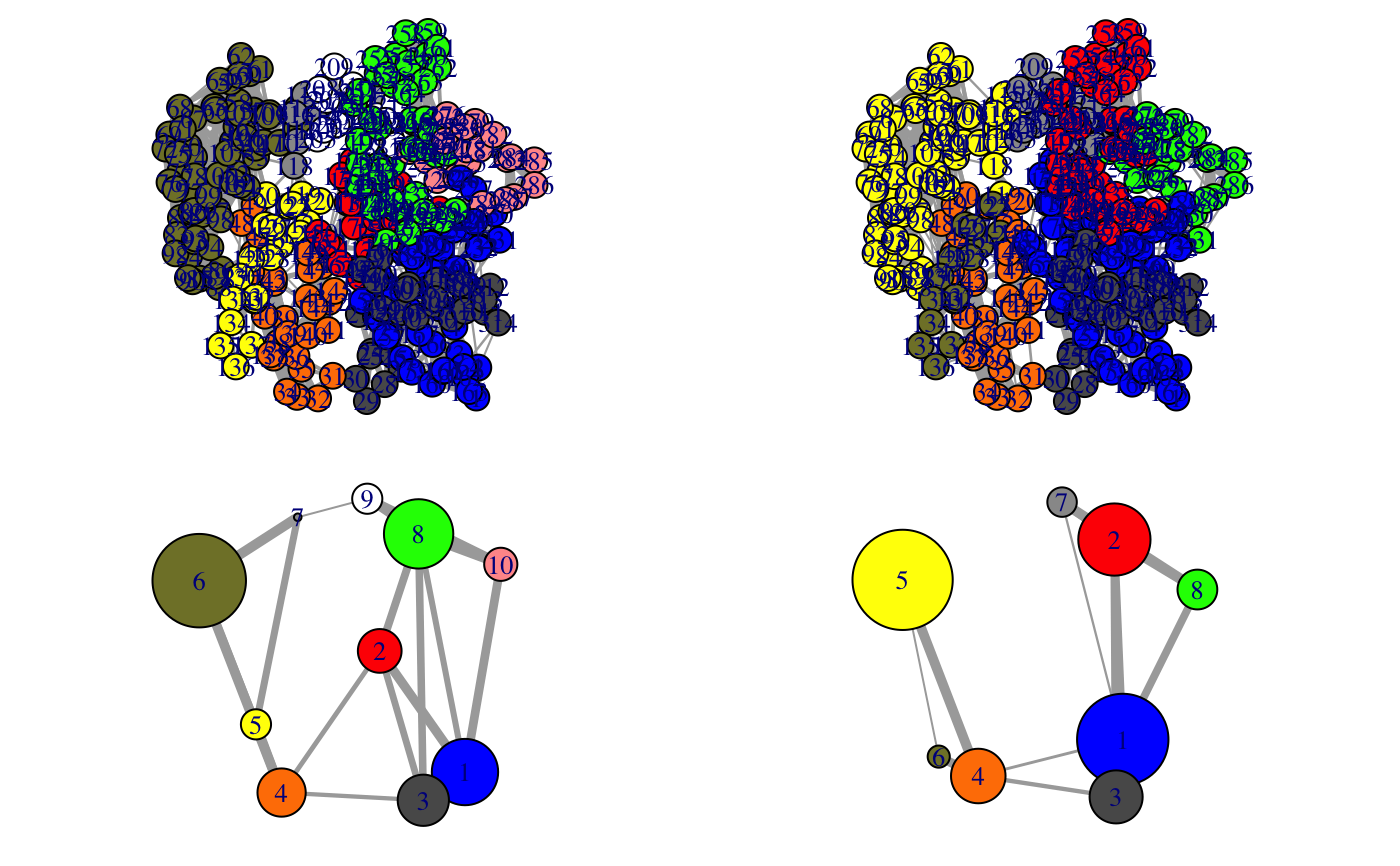
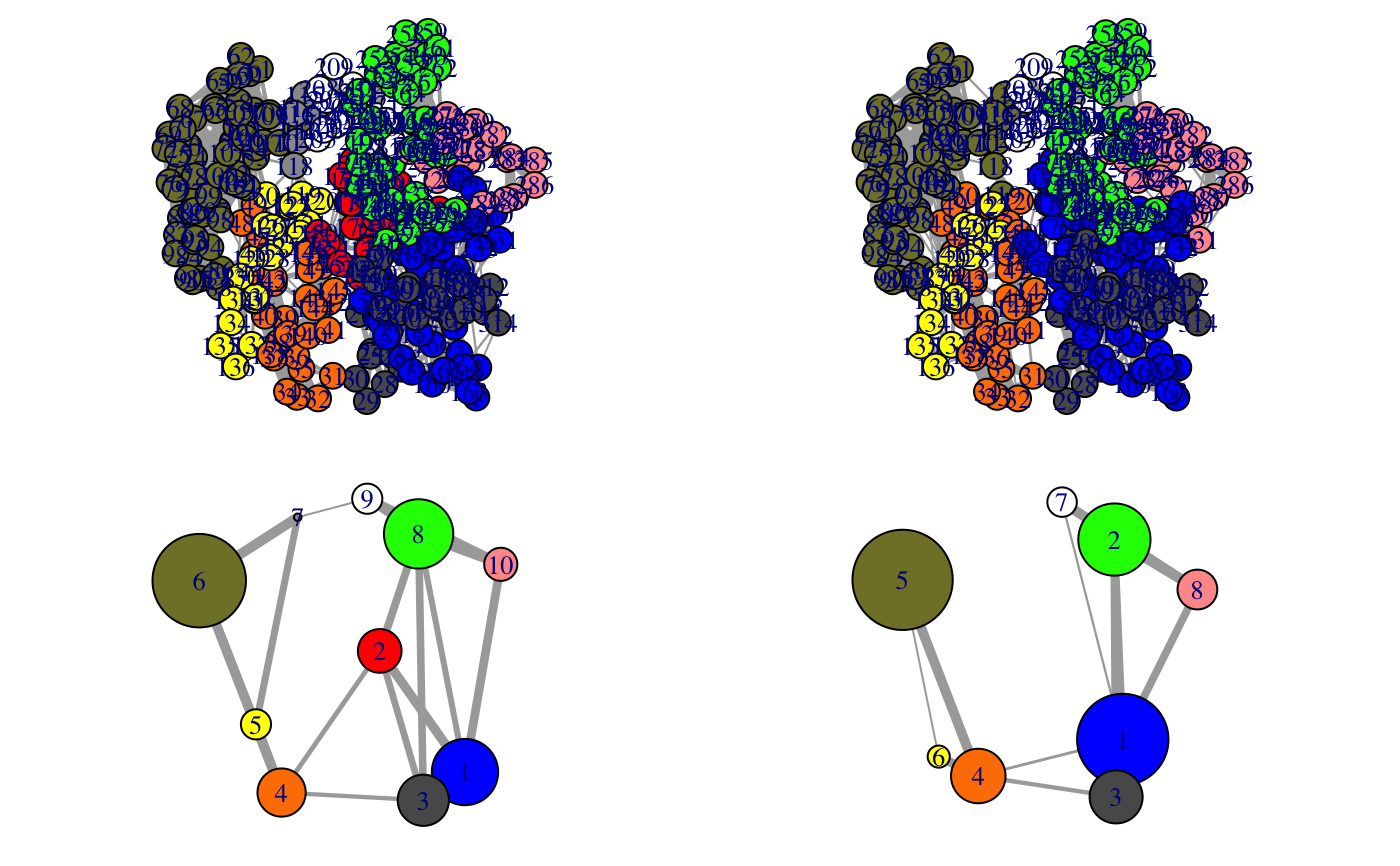
Align communities from two or more networks
community.aln.RdFind equivalent communities from two or more networks and re-assign colors to them in a consistent way across networks. A ‘new.membership’ vector is also generated for each network, which maps nodes to community IDs that are renumbered according to the community equivalency.
community.aln(x, ..., aln = NULL)
Arguments
| x, ... | two or more objects of class |
|---|---|
| aln | alignment for comparing networks with different numbers of nodes. |
Value
Returns a list of updated cna objects.
Details
This function facilitates the inspection on the variance of the community
partition in a group of similar networks. The original community numbering
(and so the colors of communities in the output of plot.cna and
vmd.cna) can be inconsistent across networks, i.e. equivalent
communities may display different colors, impeding network comparison.
The function calculates the dissimilarity between all communities and
clusters communities with ‘hclust’ funciton. In each cluster, 0 or
1 community per network is included. The color attribute of communities is
then re-assigned according to the clusters through all networks. In addition,
a ‘new.membership’ vector is generated for each network, which mapps
nodes to new community IDs that are numbered consistently across networks.
See also
cna, plot.cna, vmd.cna
Examples
# \donttest{ # Needs MUSCLE installed - testing excluded if(check.utility("muscle")) { if (!requireNamespace("igraph", quietly = TRUE)) { message('Need igraph installed to run this example') } else { ## Fetch PDB files and split to chain A only PDB files ids <- c("1tnd_A", "1tag_A") files <- get.pdb(ids, split = TRUE, path = tempdir()) ## Sequence Alignement pdbs <- pdbaln(files, outfile = tempfile()) ## Normal mode analysis on aligned data modes <- nma(pdbs, rm.gaps=TRUE) ## Dynamic Cross Correlation Matrix cijs <- dccm(modes)$all.dccm ## Correlation Network nets <- cna(cijs, cutoff.cij=0.3) ## Align network communities nets.aln <- community.aln(nets) ## plot all-residue and coarse-grained (community) networks pdb <- pdbs2pdb(pdbs, inds=1, rm.gaps=TRUE)[[1]] op <- par(no.readonly=TRUE) # before alignment par(mar=c(0.1, 0.1, 0.1, 0.1), mfrow=c(2,2)) invisible( lapply(nets, function(x) plot(x, layout=layout.cna(x, pdb=pdb, k=3, full=TRUE)[, 1:2], full=TRUE)) ) invisible( lapply(nets, function(x) plot(x, layout=layout.cna(x, pdb=pdb, k=3)[, 1:2])) ) # after alignment par(mar=c(0.1, 0.1, 0.1, 0.1), mfrow=c(2,2)) invisible( lapply(nets.aln, function(x) plot(x, layout=layout.cna(x, pdb=pdb, k=3, full=TRUE)[, 1:2], full=TRUE)) ) invisible( lapply(nets.aln, function(x) plot(x, layout=layout.cna(x, pdb=pdb, k=3)[, 1:2])) ) par(op) } }#> Warning: /var/folders/xf/qznxnpf91vb1wm4xwgnbt0xr0000gn/T//Rtmp4WslmZ/1tnd.pdb exists. Skipping download#> | | | 0% | |=================================== | 50% | |======================================================================| 100% #> Reading PDB files: #> /var/folders/xf/qznxnpf91vb1wm4xwgnbt0xr0000gn/T//Rtmp4WslmZ/split_chain/1tnd_A.pdb #> /var/folders/xf/qznxnpf91vb1wm4xwgnbt0xr0000gn/T//Rtmp4WslmZ/split_chain/1tag_A.pdb #> .. #> #> Extracting sequences #> #> pdb/seq: 1 name: /var/folders/xf/qznxnpf91vb1wm4xwgnbt0xr0000gn/T//Rtmp4WslmZ/split_chain/1tnd_A.pdb #> pdb/seq: 2 name: /var/folders/xf/qznxnpf91vb1wm4xwgnbt0xr0000gn/T//Rtmp4WslmZ/split_chain/1tag_A.pdb #> #> Details of Scheduled Calculation: #> ... 2 input structures #> ... storing 936 eigenvectors for each structure #> ... dimension of x$U.subspace: ( 942x936x2 ) #> ... coordinate superposition prior to NM calculation #> ... aligned eigenvectors (gap containing positions removed) #> ... estimated memory usage of final 'eNMA' object: 13.5 Mb #> #> | | | 0% | |=================================== | 50% | |======================================================================| 100%# }