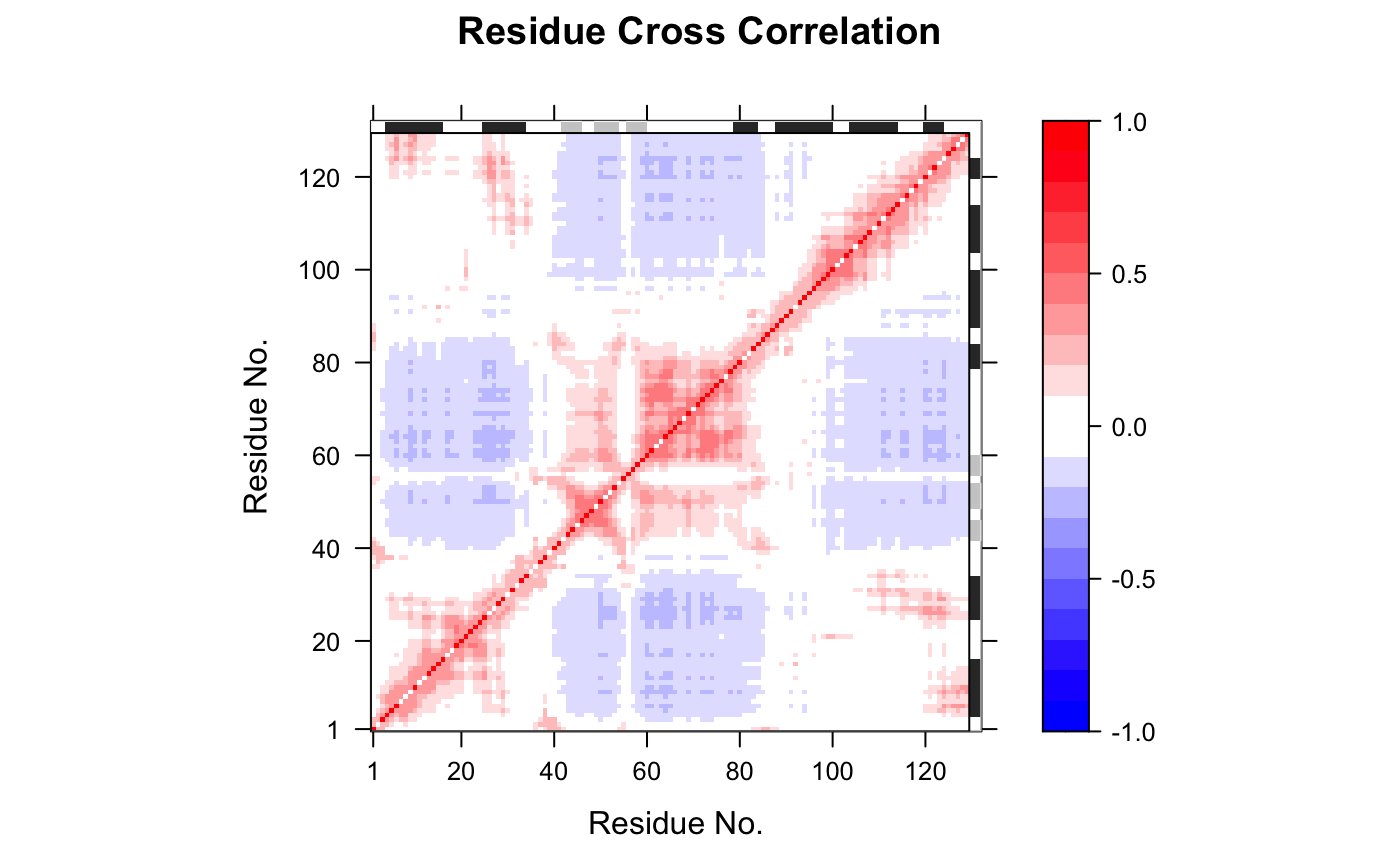
Dynamic Cross-Correlation from Gaussian Network Model
dccm.gnm.RdCalculate the cross-correlation matrix from Gaussian network model normal modes analysis.
# S3 method for gnm dccm(x, ...) # S3 method for egnm dccm(x, ...)
Arguments
| x | an object of class ‘gnm’ or ‘egnm’ as obtained from
|
|---|---|
| ... | additional arguments (currently ignored). |
Value
Returns a cross-correlation matrix.
Details
This function calculates the cross-correlation matrix from Gaussian network
model (GNM) normal modes analysis (NMA) obtained from gnm. It returns
a matrix of residue-wise cross-correlations whose elements, Cij, may be
displayed in a graphical representation frequently termed a dynamical
cross-correlation map, or DCCM. (See more details in help(dccm.nma)).
Author
Xin-Qiu Yao & Lars Skjaerven
References
Bahar, I. et al. (1997) Folding Des. 2, 173.
See also
gnm, dccm.nma, dccm.enma,
plot.dccm.
Examples
## Fetch stucture pdb <- read.pdb( system.file("examples/1hel.pdb", package="bio3d") ) ## Calculate normal modes modes <- gnm(pdb) ## Calculate correlation matrix cm <- dccm(modes) ## Plot correlation map plot(cm, sse = pdb, contour = FALSE, col.regions = bwr.colors(20), at = seq(-1, 1, 0.1))