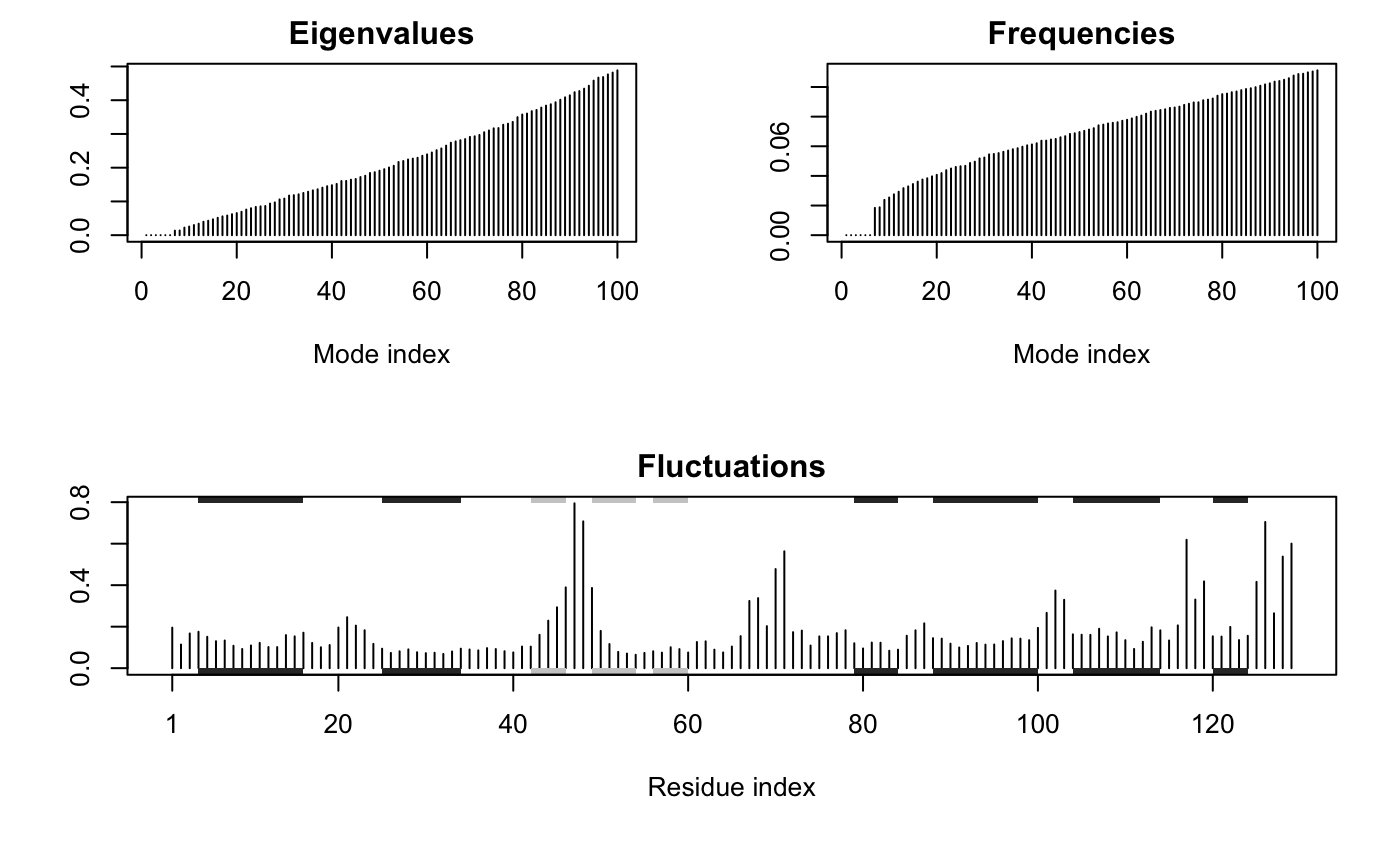
Plot NMA Results
plot.nma.RdProduces eigenvalue/frequency spectrum plots and an atomic fluctuations plot.
# S3 method for nma plot(x, pch = 16, col = par("col"), cex=0.8, mar=c(6, 4, 2, 2),...)
Arguments
| x | the results of normal modes analysis obtained with
|
|---|---|
| pch | a vector of plotting characters or symbols: see |
| col | a character vector of plotting colors. |
| cex | a numerical single element vector giving the amount by which plotting text and symbols should be magnified relative to the default. |
| mar | A numerical vector of the form c(bottom, left, top, right) which gives the number of lines of margin to be specified on the four sides of the plot. |
| ... | extra plotting arguments passed to |
Details
plot.nma produces an eigenvalue (or frequency) spectrum plot
together with a plot of the atomic fluctuations.
Value
Called for its effect.
References
Skjaerven, L. et al. (2014) BMC Bioinformatics 15, 399. Grant, B.J. et al. (2006) Bioinformatics 22, 2695--2696.
Author
Lars Skjaerven
See also
nma, plot.bio3d
Examples
## Fetch structure pdb <- read.pdb( system.file("examples/1hel.pdb", package="bio3d") ) ## Calculate modes modes <- nma(pdb)#> Building Hessian... Done in 0.01 seconds. #> Diagonalizing Hessian... Done in 0.08 seconds.